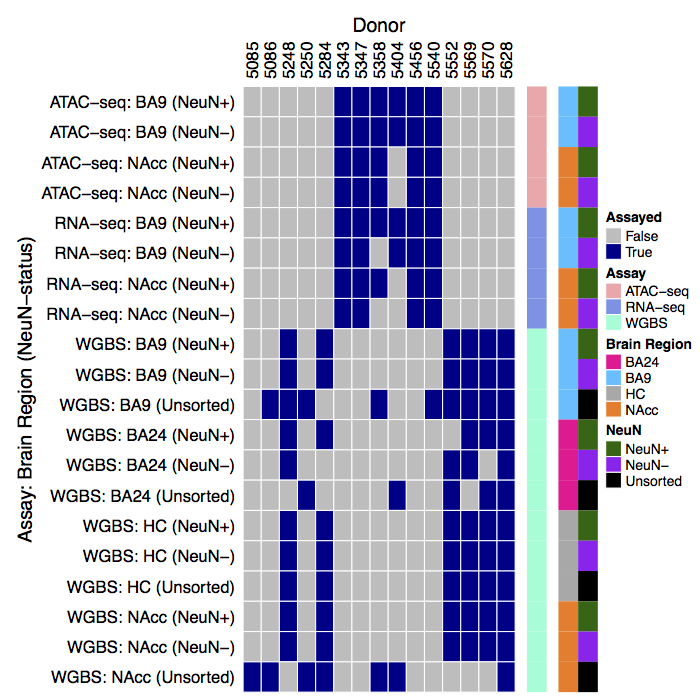
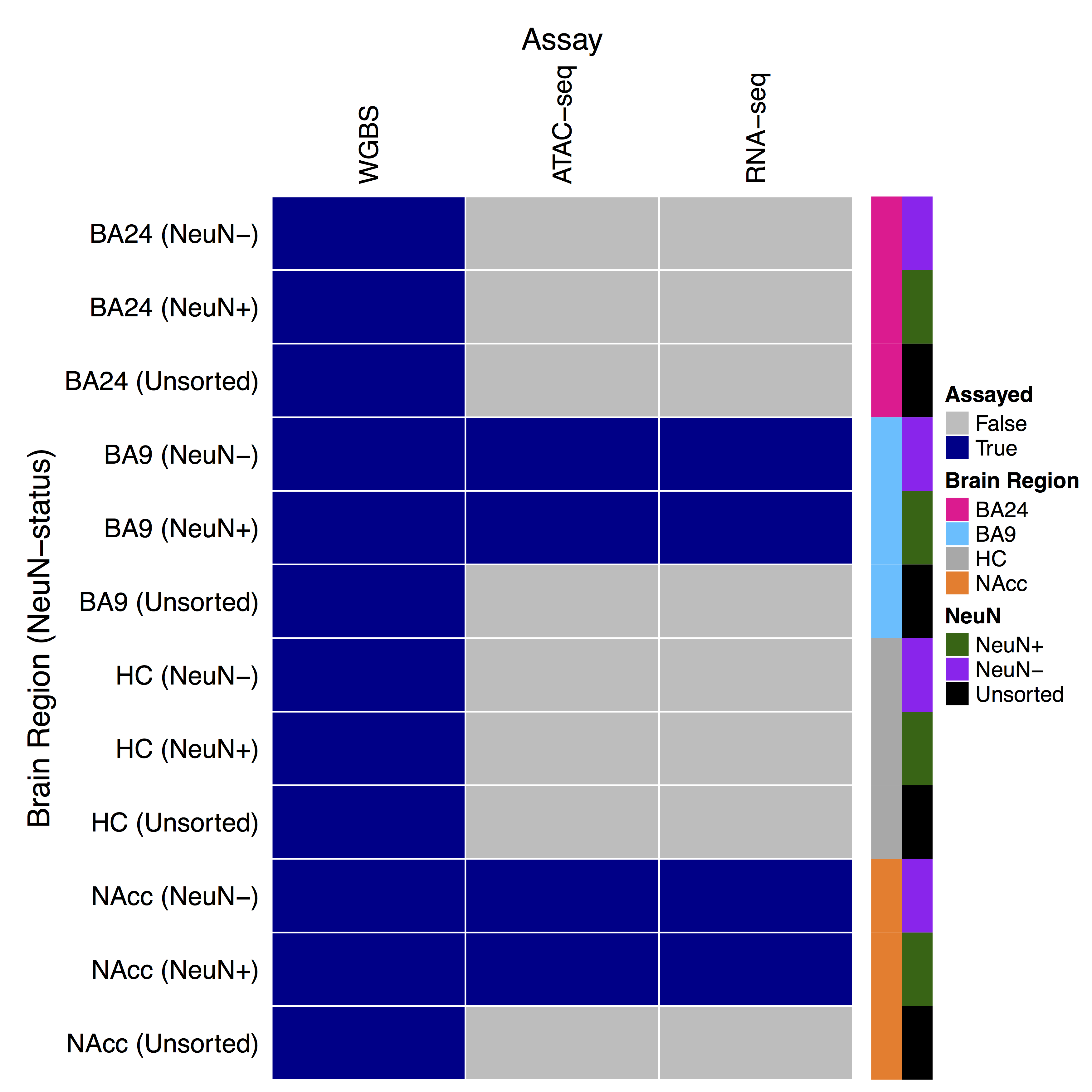
These tracks show results from genome-wide analyses of DNA methylation, chromatin accessibility, and gene expression data from 15 human brain donors [1].
We performed:
Data come from 4 brain regions:
and 2 ‘cell types’ and bulk tissue:

Methods for each assay are described in the following documents:
Please refer to [1] for further details.
Methylation and coverage tracks do not incorporate biological variability. Visually inferring regions of differential methylation, differential accessibility, or differential expression from these tracks alone is not recommended. Instead, information across all samples should be used to identify these regions and analysed with statistical methods that account for biological variability (see ‘Data Access’, below).
Raw and processed data generated during this study will be made available on the Gene Expression Omnibus (GSE96615) upon publication. We recommend that these data are used for any downstream statistical analyses.
The raw data for these tracks can be explored interactively using the Table Browser or Data Integrator. The tables can also be downloaded from our downloads server for local processing; the exact filenames can be found in the track configuration file under the bigDataUrl field. To download a track, append the bigDataUrl to the base URL of https://s3.us-east-2.amazonaws.com/brainepigenome/hg19/ For example, the track ‘Average mCG (small smooth) in BA9 (NeuN+) samples’ has bigDataUrl of BA9_pos.small_smooth.mCG.bw and can be downloaded from https://s3.us-east-2.amazonaws.com/brainepigenome/hg19/BA9_pos.small_smooth.mCG.bw.
Questions should be directed to Peter Hickey.